
Actas del Congreso Nacional de
Tecnología Aplicada a Ciencias
de la Salud



Actas del Congreso Nacional de Tecnología Aplicada a Ciencias de la Salud Vol. 4, 2022
In this work, we introduce the first version of an algorithm to obtain SARS-CoV-2 cells 3D profiles by means of data integration in images obtained in a clinical tomographic study in rodents’ brain tissue with bioluminescence. We propose an algorithm based on Legendre polynomials along with a phase recovery isomorphism to be applied in SARS-CoV-2 cells (virion) morphologic analysis. We show the results obtained from the analysis of a previous study in infected tissue by a SARS-CoV-2 virus cell culture from microscopy and tomography. We analyze the corresponding images by carrying out a segmentation procedure (SARS-CoV-2 cells) by applying a new filtering algorithm in the Fourier space. We show our final results in a numerical integration covering the surface interpreting the spikes in Blender.
Keywords: SARS-CoV-2, 3D Model, ROI Segmented, Live Bioluminescence, Medical Imaging
Presentamos una primera versión de un algoritmo capaz de obtener los perfiles 3D de células SARS-CoV-2 al integrar los datos contenidos en imágenes obtenidas de un estudio clínico de tomografía sobre tejido cerebral de roedores en un estudio de bioluminiscencia. Proponemos un algoritmo basado en polinomios de Legendre y un isomorfismo de recuperación de fase para ser aplicado en un análisis morfológico de células SARS-CoV-2 (virión). Los resultados se obtienen del análisis de un estudio previo en tejido infectado por un cultivo celular del virus SARS-CoV-2, obtenido de microscopía y tomografía. Analizamos dichas imágenes realizando un procedimiento de segmentación (células SARS-CoV-2) aplicando un nuevo algoritmo de filtrado en el espacio de Fourier mostrando nuestros resultados finales en una integracion numerica cubriendo dicha superficie que interpreta las espiculas, en Blender.
Palabras Clave: SARS-CoV-2, Modelo 3D, ROI Segmentada, Bioluminiscencia in Vivo, Imágenes Médicas
Computer vision, profilometry, and 3D digitization of objects are tools that merge several techniques, constituting 3D visualization and object digital construction [1, 2, 3, 4]. These techniques have grown in lockstep with the technological innovation in computational hardware and software development [3, 5], as can be seen in some fields in sciences such as medicine, biology, chemistry, and 3D modeling-based studies as numerical approximations [6]. On the other hand, such techniques allow us to increase the resolution in visual analysis, useful in the diagnosis [3, 4, 7, 8]. The latter marked a milestone in the study and recognition of anomalous and irregular patterns in the numerical analysis of raw data (without prior treatment), interpreted as an increase in the resolution, allowing us to observe and recognize such irregularities not distinguished at first sight easily. Such developments in 3D vision have applications in biology, chemistry, medicine [4, 7], architecture, engineering, etc.
Regarding clinical tests, these techniques marked a benchmark in the analysis of biological samples to identify anomalies, and pathologies (cysts, tumors, calcifications, cancer, etc). These techniques allow us to perform analysis without contaminating the samples and avoid additional studies in several cases (considering that additional studies are a useful tool to validate the results from previous studies), considering processing techniques and 3D vision with no contact [1, 7, 9]. The latter is fundamental in studies with high-risk agents, as in several clinical, biological, and chemical studies keeping the sample intact and avoiding contamination. In this case, the pathological agents are a technique that reduces the leaks and contagion risks to extend the analysis in the data recovery.
Hence, in this work, we obtain 3D profiles of SARS-CoV-2 cells in rodents, observed in brain tomography, and analyzed in a live bioluminescence imaging study (BLI) by Ullah et al. [10]. In the work by Ullah et al. [10], a live Bioluminescence Imaging (BLI) is performed to follow up in real time a Neutralizing Antibodies treatment as a therapy in rodents K18-hACE2 under a SARS-CoV-2 nanoluciferase (SARS-CoV-2-N) cells culture infected intranasally. From such a study, 3D tomographic images (EM tomography) are obtained from rodents’ brain tissue with a SARS-CoV-2 culture. Such a tomographic technique is often called 3D tomographic reconstruction [11, 12].
Thus, we thoroughly recall and study the work by Ullah et al. [10], where type 18-hACE2 rodents (genetically modified rodents for studies related to the current COVID-19 pandemic) are infected with SARS-CoV-2 intra-nose. After inoculation, the infection of the SARS-CoV-2 in rodents was validated to determine the virus propagation in the tissue using the BLI technique. The author presents such results as well as some proposals to stop the propagation. However, from all the results, we focus on brain tomography images, where a large population of SARS-CoV-2 cells with considerable growth, can be observed extensively, along with their corresponding videos. The results in Ullah et al. [10] are based on NAbs cells (Neutralizing antibodies), studied as immunologic protection from antibody culture (requiring additional studies and analysis since they need to be validated in humans and other rodent species) in the infected rodents, to perform an analysis using live bioluminescence imaging (BLI). Subsequently, from the obtained BLI data, we analyze the images to determine certain selection criteria depending on the SARS-CoV-2 propagation to determine the organ to be studied, and hence, we identified the infected zones in the ROI (regions of interest [7]), lungs, brains, and testicles, to subsequently conduct directed histology studies and electronic tomography [10]. The latter revealed that SARS-CoV-2 is related to the lungs' capillary endothelial cells and alveolar macrophage cells. Similarly, in the testicles, a large population of virions was observed inside the pleomorphic membranes, in the brain, in brain cells, and inner parts of the dendrites. We focus on the brain ROI, included as additional material Visualization 1 (Video S2 [10])1 , where we observe SARS-CoV-2 cells in low contrast. In the same tomography, we notice several planes on the brain tissue, giving place to different squares per time unit in the video. We propose the application of these computer vision techniques to perform the numerical integration of the data shown in the video to subsequently build the 3D model from the tomography data, implying a new technique that can be included as an integration tool and 3D recognition. Our proposal uses the 2D-LP (two-dimensional Legendre polynomials) interpolator and a modified Gerchberg-Saxton code version with the Legendre Polynomials, which we presented and applied in Arriaga et al. [3, 4], and we applied to the frames obtained as captures from Visualization 1.
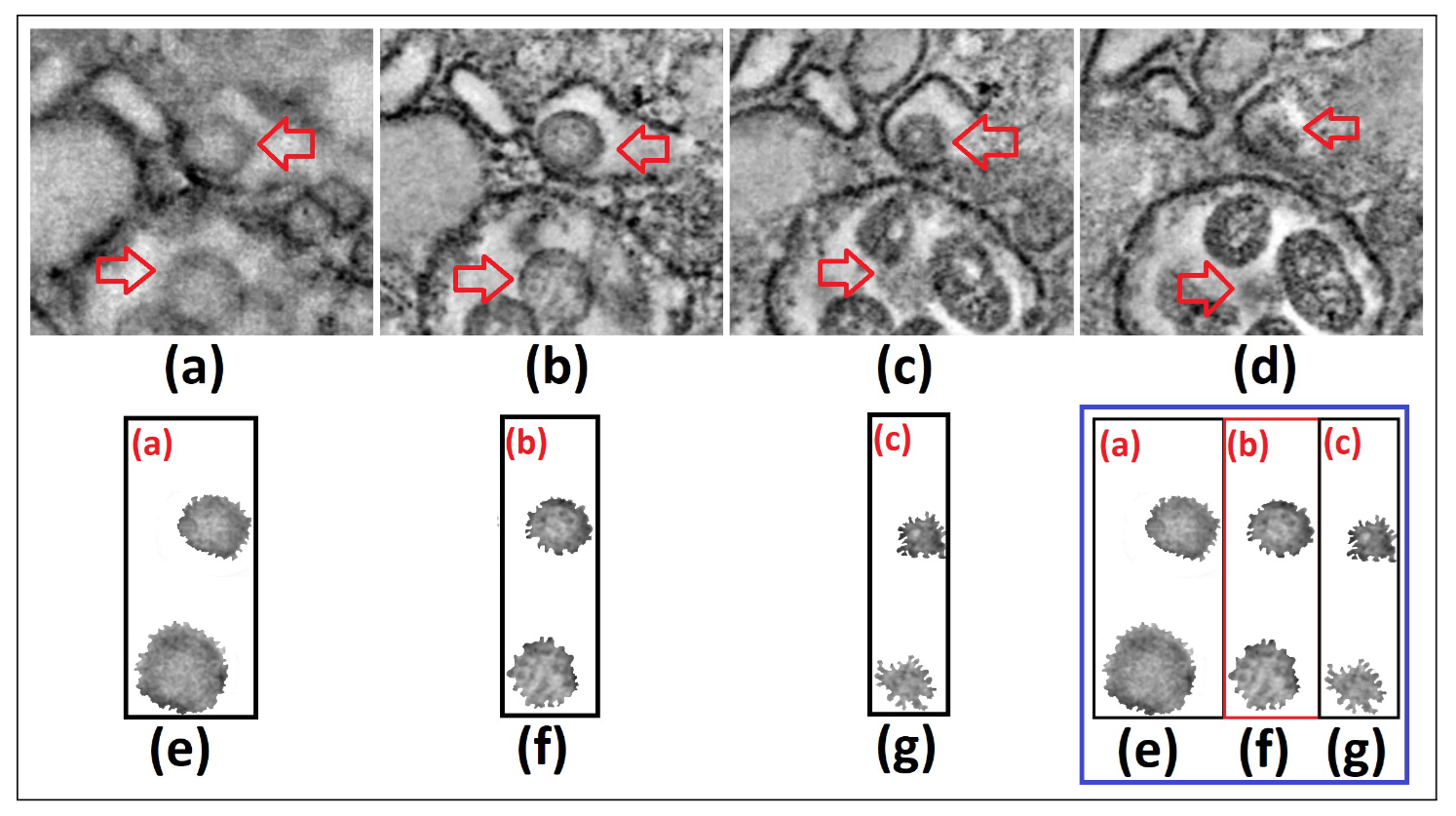
Figure 1. We show 4 captures or frames from Visualization 1 [10] around t=32 sec (a)-(d), and the segmentation and gray tones mapping of the SARS-CoV-2 cell
To obtain our results, through 3D data integration, we extract 4 different frames from Visualization 1, close to 32 seconds. We subsequently segment the image in the upper-right frame and we select from it the SARS-CoV-2 cells for analysis. We perform a subsequent and more refined segmentation procedure in terms of intensity heights (with the SARS-CoV-2 cells having darker tones) and later in gray tones [0,255], to obtain the information of such cells only [Fig. 1 (a)-(d)], resembling transparency only with gray tones different from 0 and 255 [Fig. 1 (e)-(g)].
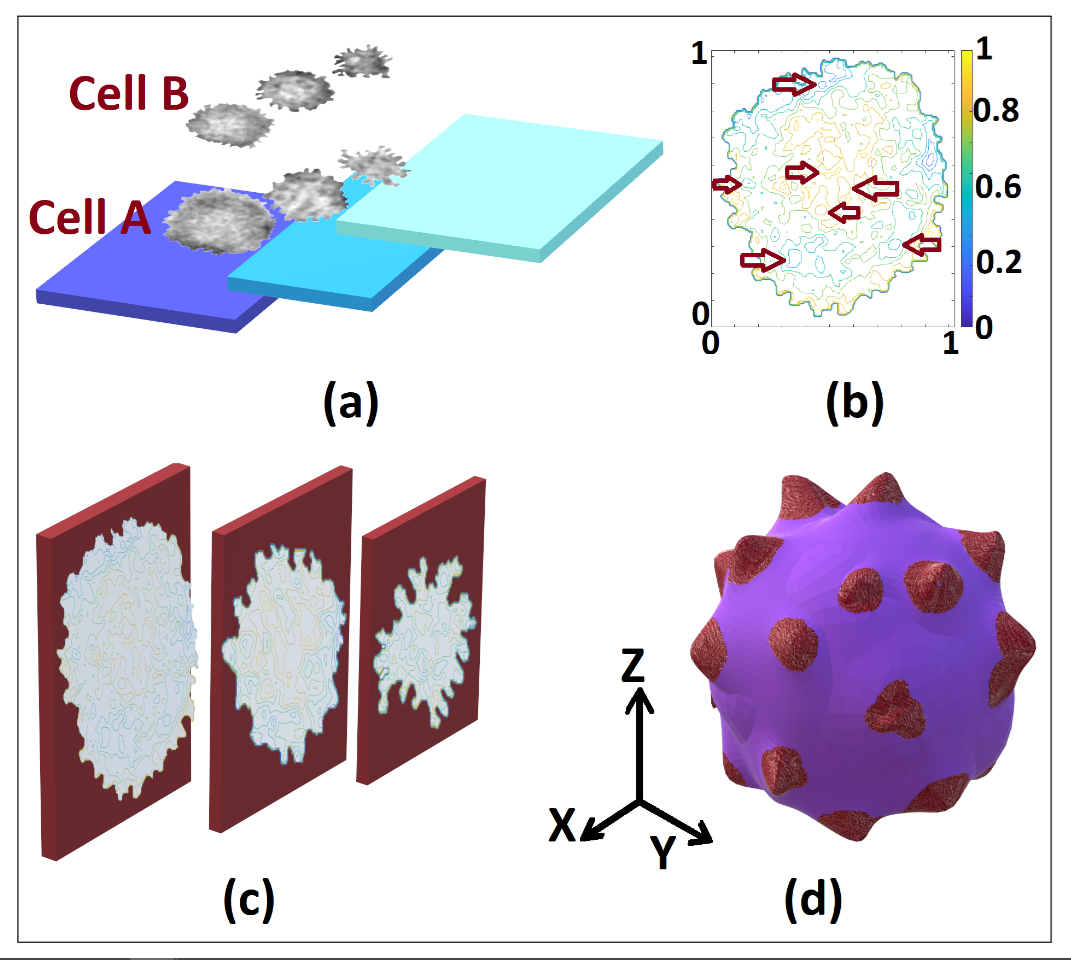
Figure 2. Levels distribution in the tomography of cells A and B (a), highlighting non-visible features in each 3D normalized matrix frame levels (b), considered to build the 3D model (c) and the resulting model (d) of the SARS-CoV-2 cell in our proposal
We highlight that the resolution of the reconstructed 3D object depends on the frames and the video quality. We obtained the results shown in Fig. 2 for two SARS-CoV-2 cells, identified in Fig. 1. In Fig. 2 (a), we show the frames selection and their location in the 3D matrix, displayed by layers or data levels. Using our numerical proposal, we obtain an intermediate theoretical frame between two experimental adjacent and disjoint frames until the size of the desired 3D matrix (512x512x512) is reached [8]. Subsequently, we integrate the data to obtain the object or its level curves according to the observed 3D matrix. In Fig. 2 (a), we bear in mind that the isophotes of each 3D level can be obtained by increasing or strengthening the study of the cell in its inner regions, recognizing the non-visible patterns in the tomography. We obtained the 3D model in Fig. 2 (d) using Paint 3D (Microsoft), artificially adding colors and contrasts to visualize the spikes since the results in Blender hindered during the generation of the 3D object with ply format for the point cloud. We found the most significant error in the results rendering given that the results were not properly integrated into the point cloud, without distinguishing the spikes. Hence, our final results are the data integration in the point cloud [13] in Matlab, which we subsequently export to Blender to finally export as a 3D Paint object, where we add false color to obtain Fig. 2 (d).
Our numerical proposal is focused on obtaining 3D numerical profiles from data interpolation, abiding by the probability and sampling theory, since we interpolate the data in a multilinear fashion to obtain intermediate frames. The latter gives us the possibility to carry out an approximate analysis of probable elements in the inner regions of the cell in terms of the tomography resolution. We emphasize that our proposal is not based on other models to obtain three-dimensionally the SARS-CoV-2 virion. Our proposal obtains it from accurate metadata instead of reconstructing each phase cell according to its selection in the proper ROI. Subsequently, the results are obtained from such cells given their morphology and other elements. Thus, the results shown in Fig. 2 (b) correspond to each SARS-CoV-2 cell studied and are not based on any a priori computational model.
acknowledgments
The authors want to thank Benemérita Universidad Autónoma de Puebla (BUAP) for the support given during this research work. We also thank CONACYT (Consejo Nacional de Ciencia y Tecnología, México).
Disclosures
The authors declare no conflict of interest.
1.- We will use the name Visualization 1 to refer to video S2 in [10] throughout the text. The images under analysis were captured as frames from Visualization 1 and shown in Fig. 1.